Publications
2025
-
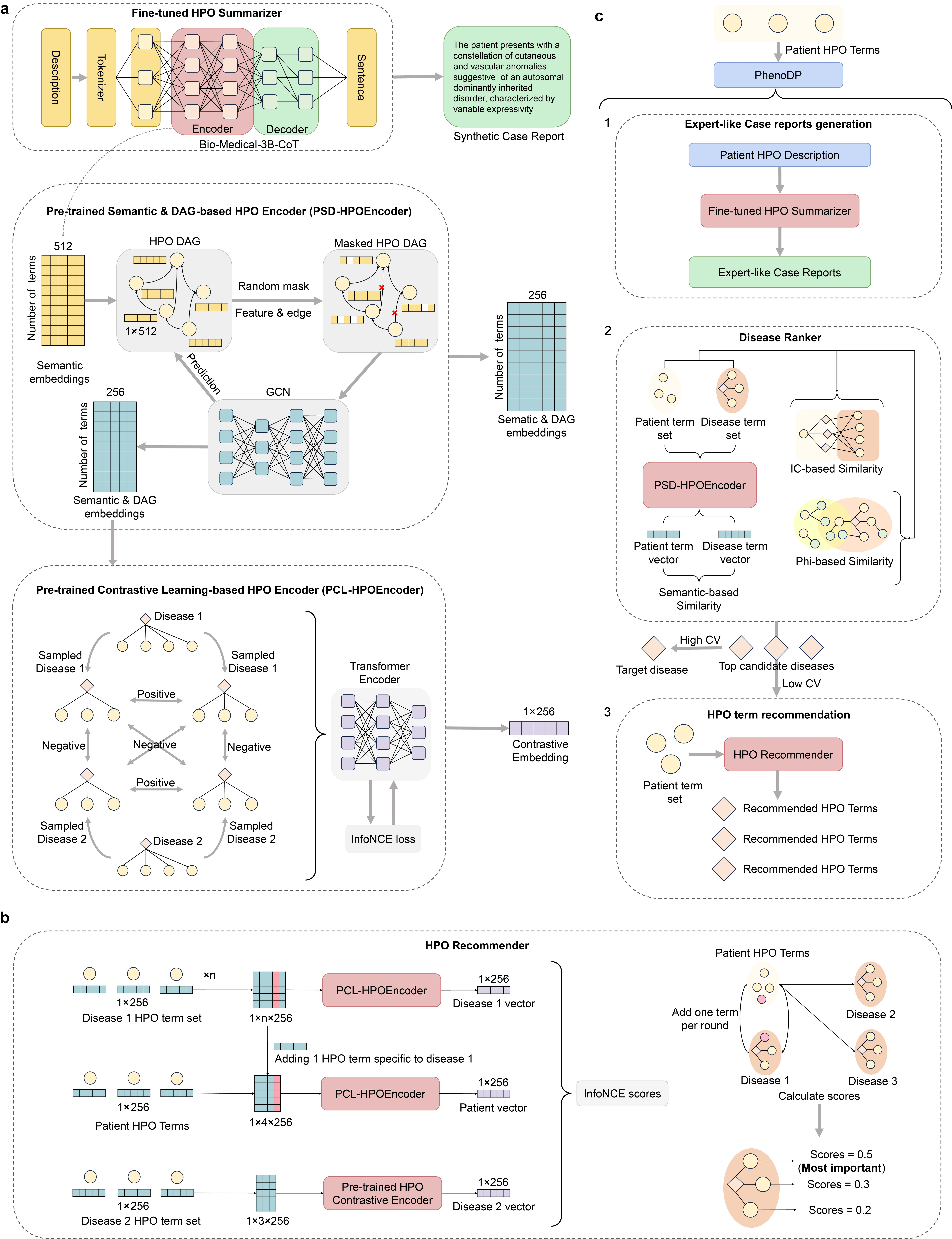 PhenoDP: leveraging deep learning for phenotype-based case reporting, disease ranking, and symptom recommendationGenome Medicine, 2025
PhenoDP: leveraging deep learning for phenotype-based case reporting, disease ranking, and symptom recommendationGenome Medicine, 2025 -
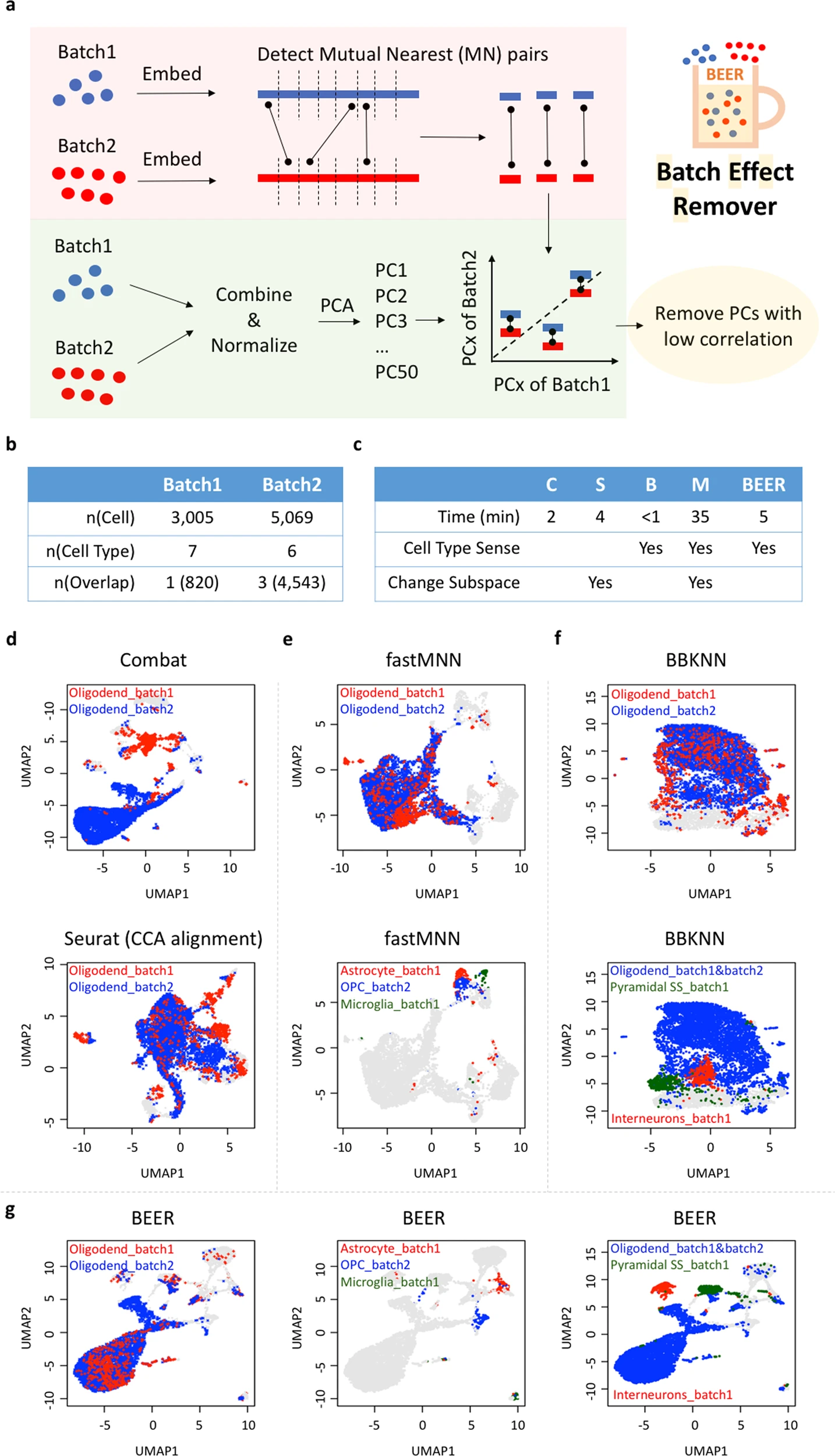
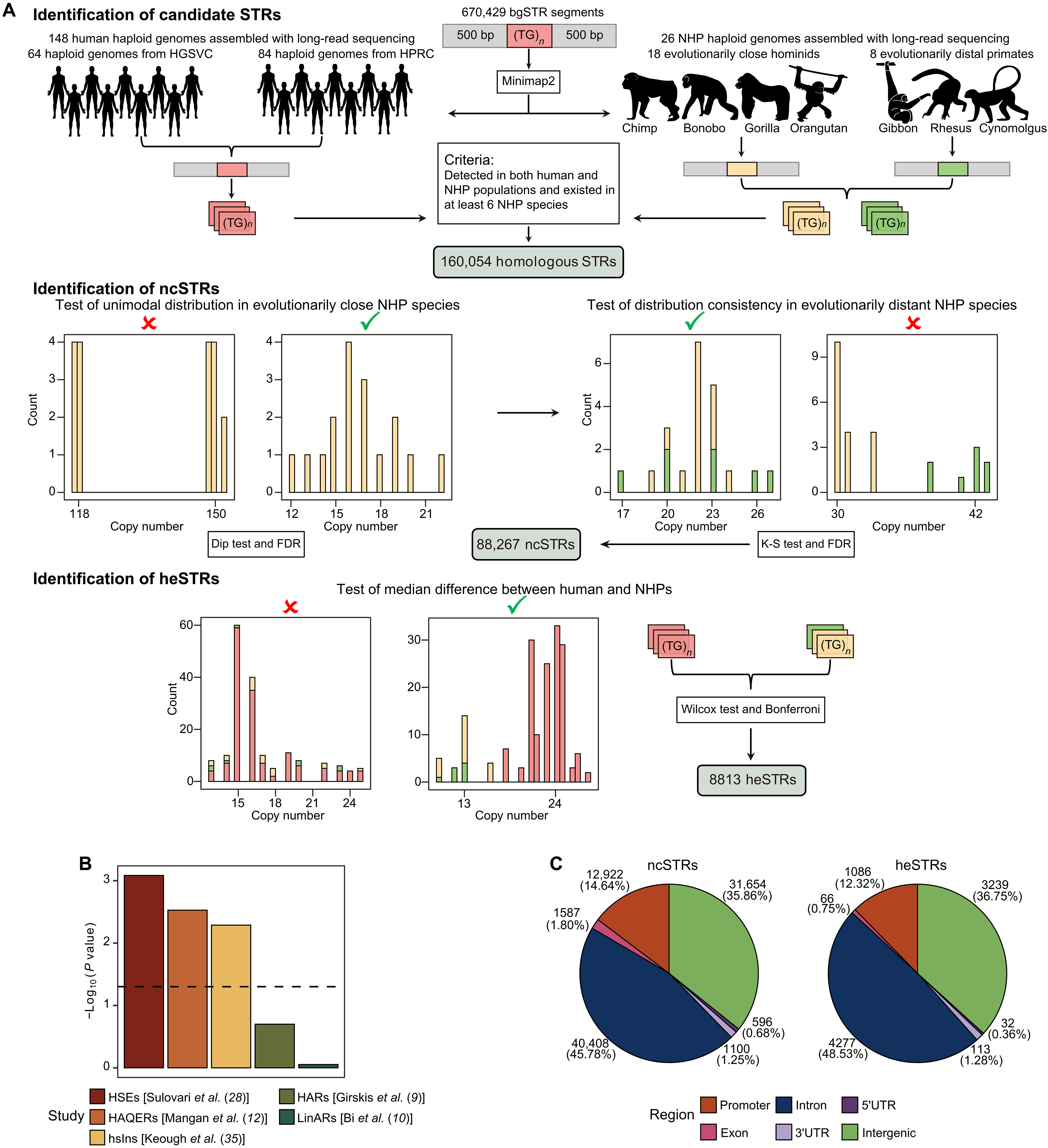 Association of human-specific expanded short tandem repeats with neuron-specific regulatory featuresScience Advances, 2025
Association of human-specific expanded short tandem repeats with neuron-specific regulatory featuresScience Advances, 2025
2024
-
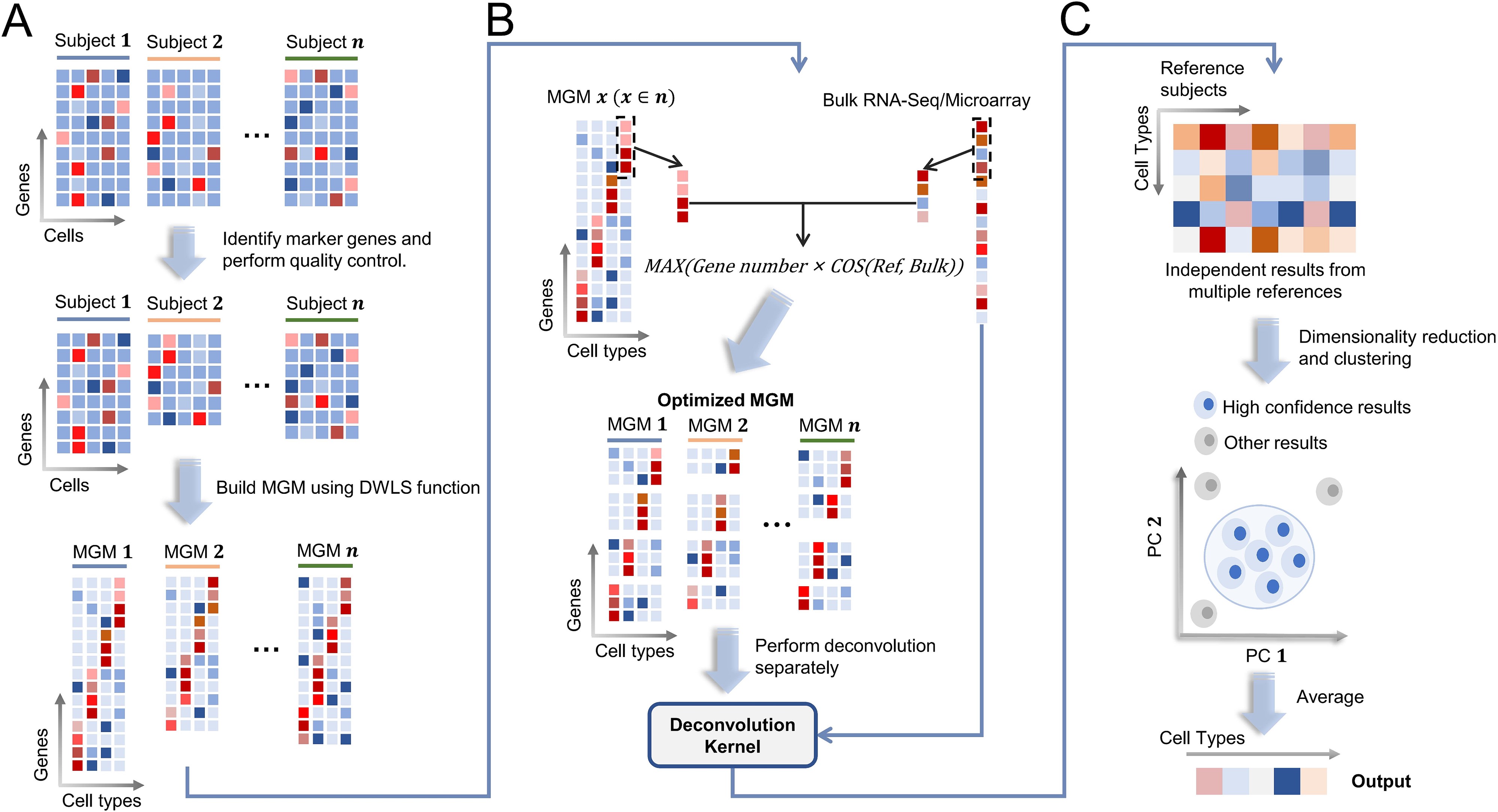 ReCIDE: robust estimation of cell type proportions by integrating single-reference-based deconvolutionsBriefings in Bioinformatics, 2024
ReCIDE: robust estimation of cell type proportions by integrating single-reference-based deconvolutionsBriefings in Bioinformatics, 2024 - MGA loss-of-function variants cause premature ovarian insufficiencyThe Journal of Clinical Investigation, 2024
2023
-
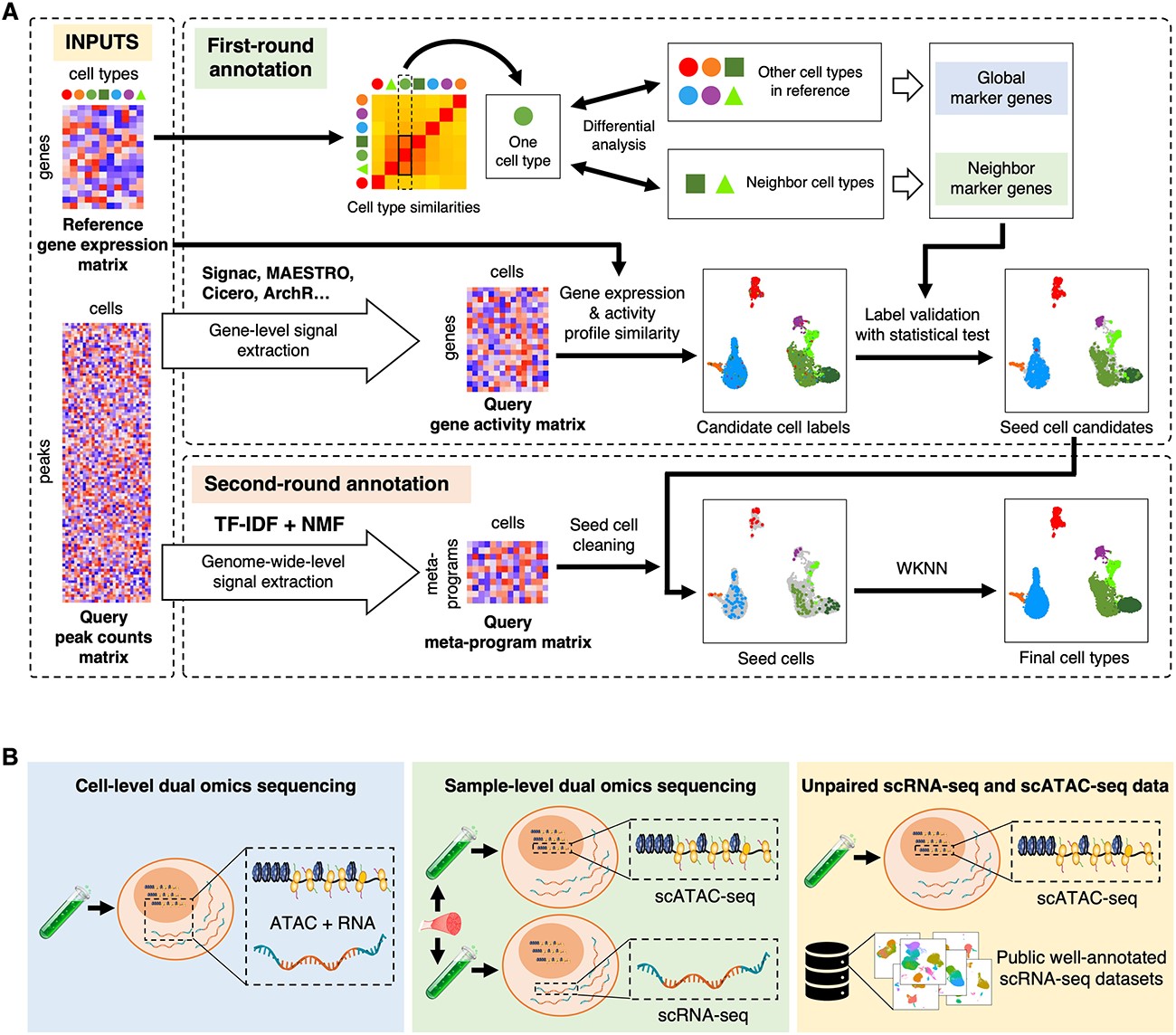 AtacAnnoR: a reference-based annotation tool for single cell ATAC-seq dataBriefings in Bioinformatics, 2023
AtacAnnoR: a reference-based annotation tool for single cell ATAC-seq dataBriefings in Bioinformatics, 2023 - Single-cell RNA-seq reveals developmental deficiencies in both the placentation and the decidualization in women with late-onset preeclampsiaFront Immunol, 2023
- Kinase-independent role of mTOR and on-/off-target effects of an mTOR kinase inhibitorLeukemia, 2023Epub 2023 Aug 2.PMID: 37532788
2022
- Deconvolution of a Large Cohort of Placental Microarray Data Reveals Clinically Distinct Subtypes of PreeclampsiaFrontiers in Bioengineering and Biotechnology, 2022
-
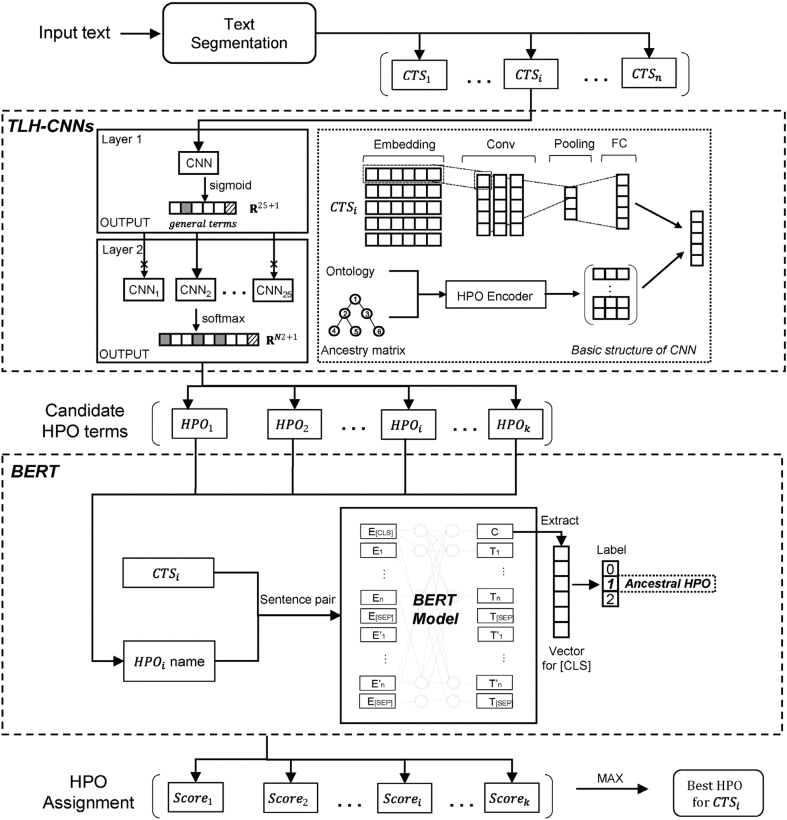 PhenoBERT: a combined deep learning method for automated recognition of human phenotype ontologyIEEE/ACM Transactions on Computational Biology and Bioinformatics, 2022
PhenoBERT: a combined deep learning method for automated recognition of human phenotype ontologyIEEE/ACM Transactions on Computational Biology and Bioinformatics, 2022 - Large-scale prediction of key dynamic interacting proteins in multiple cancersInternational journal of biological macromolecules, 2022
- MUC1 triggers lineage plasticity of Her2 positive mammary tumorsOncogene, 2022
- Large-scale prediction of key dynamic interacting proteins in multiple cancers Int J Biol Macromol2022
- Deconvolution of a Large Cohort of Placental Microarray Data Reveals Clinically Distinct Subtypes of PreeclampsiaFront Bioeng Biotechnol. Jul, 2022
- scMAGIC: accurately annotating single cells using two rounds of reference-based classificationNucleic Acids Research, 2022
- PhenoBERT: a combined deep learning method for automated recognition of human phenotype ontologyIEEE/ACM Trans Comput Biol Bioinform, 2022
-
- Single‐cell transcriptomics reveals pathogenic dysregulation of previously unrecognised chondral stem/progenitor cells in children with microtiaClin Transl Med, 2022
2021
- CRMarker: A manually curated comprehensive resource of cancer RNA markersInt J Biol Macromol, 2021
- Adaptive responses to mTOR gene targeting in hematopoietic stem cells reveal a proliferative mechanism evasive to mTOR inhibitionProc Natl Acad Sci U S A, 2021PMID: 33443202; PMCID: PMC7817152.
2020
-
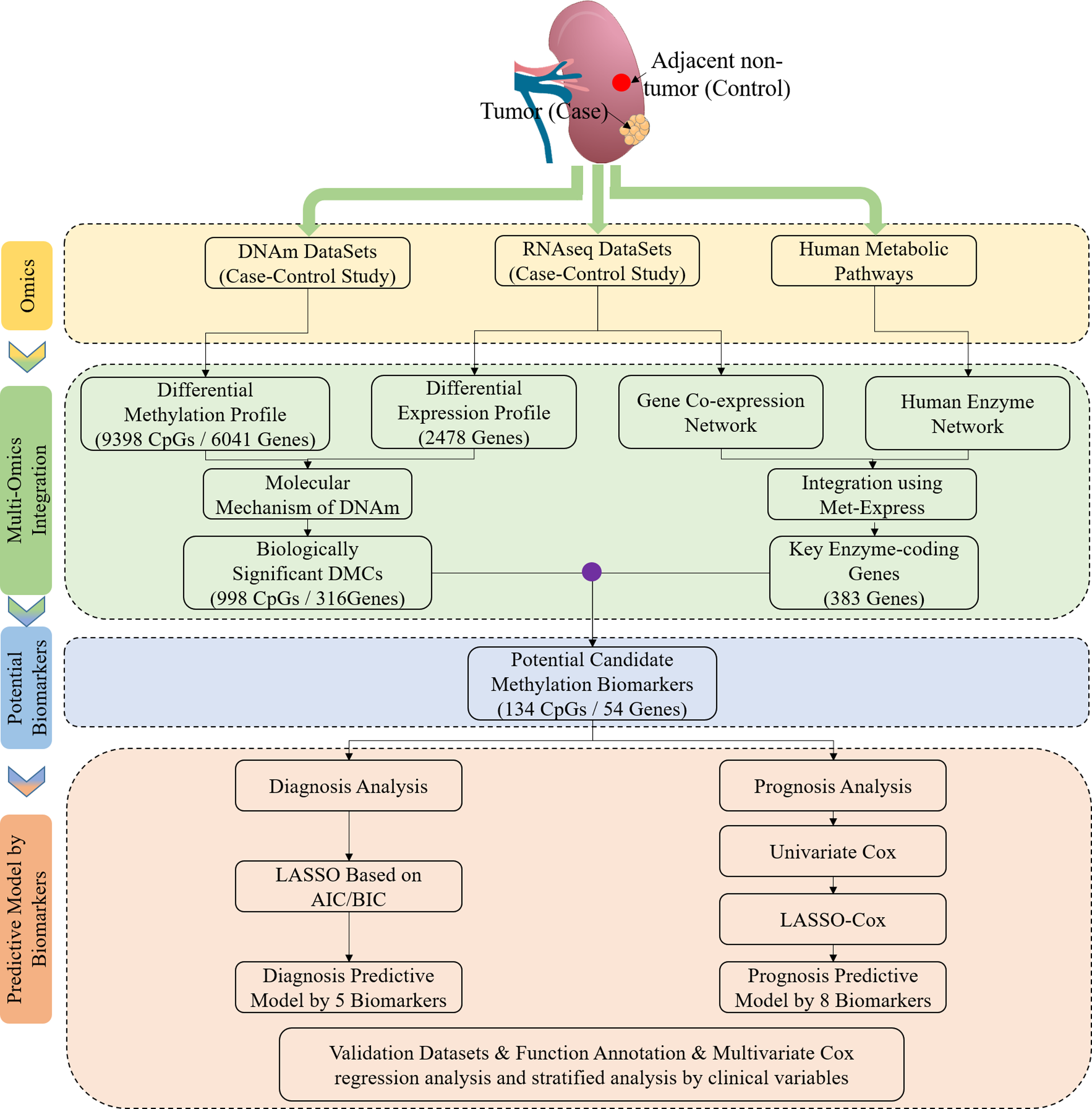 Identification of DNA methylation patterns and biomarkers for clear-cell renal cell carcinoma by multi-omics data analysisPeerJ, 2020
Identification of DNA methylation patterns and biomarkers for clear-cell renal cell carcinoma by multi-omics data analysisPeerJ, 2020 - Global synthesis of effects of plant species diversity on trophic groups and interactionsNat Plants, 2020Epub 2020 May 4.
- Unsupervised Inference of Developmental Directions for Single Cells Using VECTORCell Reports, 2020PMID: 32846127
- Identification of DNA methylation patterns and biomarkers for clear-cell renal cell carcinoma by multi-omics data analysisPeer J, 2020
- Network Properties of Cancer Prognostic Gene Signatures in the Human Protein InteractomeGenes, 2020
-
2019
-
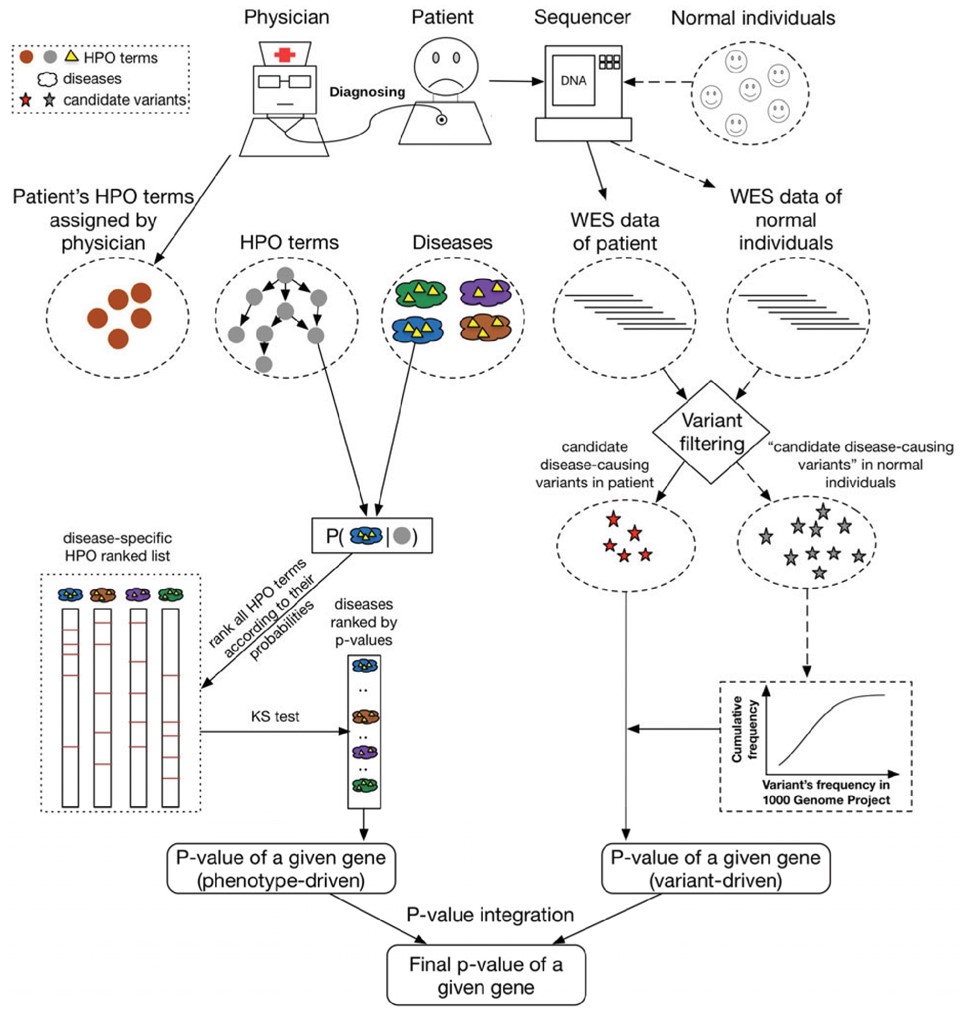 PhenoPro: a novel toolkit for assisting in the diagnosis of Mendelian diseaseBioinformatics, 2019
PhenoPro: a novel toolkit for assisting in the diagnosis of Mendelian diseaseBioinformatics, 2019 - Single-Cell Transcriptomics in Medulloblastoma Reveals Tumor-Initiating Progenitors and Oncogenic Cascades during Tumorigenesis and RelapseCancer Cell, 2019pii: S1535-6108(19)30336-8.
- Evolution and Comprehensive Analysis of DNaseI Hypersensitive Sites in Brain-Related Genes Regulatory Regions of PrimatesFrontiers in Genetics, 2019eCollection 2019. (PMID: 30930929
2018
- Increasing plant diversity with border crops reduces insecticide use and increases crop yield in urban agricultureElife, 2018pii: e35103.
2017
- Dynamic Editome of Zebrafish under Aminoglycosides Treatment and Its Potential Involvement in OtotoxicityFront Pharmacol. Nov, 2017eCollection 2017. (PMID: 29213239
- SPRINT: an SNP-free toolkit for identifying RNA editing sitesBioinformatics, 2017PMID: 29036410
- Whole genome sequencing identifies a novel ALMS1 gene mutation in two Chinese siblings with Alström syndromeBMC Med Genet. Jul, 2017PMID: 28724398
- GenePANDA-a novel network-based gene prioritizing tool for complex diseasesScientific Reports, 2017PMID: 28252032
- Regulation of mRNA splicing by MeCP2 via epigenetic modifications in the brainScientific Reports, 2017PMID: 28211484
2016
- China’s community-based strategy of universal preconception care in rural areas at a population level using a novel risk classification system for stratifying couples preconception health statusBMC Health Serv Res, 2016PMID: 28031048
- Cellulose synthesis genes CESA6 and CSI1 are important for salt stress tolerance in ArabidopsisJ Integrative Plant Biology, 2016PMID: 26503768
- Prediction of Candiyear Drugs for Treating Pancreatic Cancer by Using a Combined ApproachPloS one, 2016PMID: 26910401
- An expanded evaluation of protein function prediction methods shows an improvement in accuracyM. J, 2016PMID: 27604469
- Explaining the disease phenotype of intergenic SNP through predicted long range regulationNucleic acids research, 2016PMID: 27280978
- Genome-Wide Identification of Regulatory Sequences Undergoing Accelerated Evolution in the Human GenomeMolecular Biology and Evolution, 2016PMID: 27401230
- GoFDR: A sequence alignment based method for predicting protein functionsMethods, 2016PMID: 26277418
- LEGO: a novel method for gene set over-representation analysis by incorporating network-based gene weightsScientific Reports, 2016PMID: 26750448
- GSA-Lightning: ultra-fast permutation-based gene set analysisBioinformatics, 2016PMID: 27296982
2015
- Positive selection on D-lactate dehydrogenases of Lactobacillus delbrueckii subspecies bulgaricusIET Systems Biology, 2015PMID: 26243834
- Prediction of Metabolic Gene Biomarkers for Neurodegenerative Disease by an Integrated Network-Based ApproachBioMed research international, 2015PMID: 26064912
- Solute Carrier Family 26 Member a2 (slc26a2) Regulates Otic Development and Hair Cell Survival in ZebrafishPloS one, 2015PMID: 26375458
2014
- microRNA expression profiling of heart tissue during fetal developmentInternational journal of molecular medicine, 2014PMID: 24604530
- Short-term and continuing stresses differentially interplay with multiple hormones to regulate plant survival and growthMolecular plant, 2014PMID: 24499771
- The effect of multiple single nucleotide polymorphisms in the folic acid pathway genes on homocysteine metabolismBioMed research international, 2014PMID: 24524080
2013
- Support vector machine: classifying and predicting mutagenicity of complex mixtures based on pollution profilesToxicology, 2013PMID: 23395826
- Revealing of Mycobacterium marinum transcriptome by RNA-seqPloS one, 2013PMID: 24098731
- A large-scale evaluation of computational protein function predictionNature methods, 2013PMID: 23353650
- Combining Hi-C data with phylogenetic correlation to predict the target genes of distal regulatory elements in human genomeNucleic acids research, 2013PMID: 24003029
- Integration of cancer gene co-expression network and metabolic network to uncover potential cancer drug targetsJournal of proteome research, 2013PMID: 23590569
- Functional-network-based gene set analysis using gene-ontologyPloS one, 2013PMID: 23418449
2012
- Epigenetic features are significantly associated with alternative splicingBMC genomics, 2012PMID: 22455468
- Pollution trees: identifying similarities among complex pollutant mixtures in water and correlating them to mutagenicityEnvironmental science & technology, 2012PMID: 22680987
- Dynamic population changes in Mycobacterium tuberculosis during acquisition and fixation of drug resistance in patientsJ Infect Dis, 2012Epub 2012 Sep 14.
- Virion protein 16 induces demethylation of DNA integrated within chromatin in a novel mammalian cell modelActa biochimica et biophysica Sinica, 2012PMID: 22120152
- SAP–a sequence mapping and analyzing program for long sequence reads alignment and accurate variants discoveryPloS one, 2012PMID: 22880129
- An iterative network partition algorithm for accurate identification of dense network modulesNucleic acids research, 2012PMID: 22121225
- 3rd et alThe Journal of infectious diseases, 2012PMID: 22984115
- Molecular mechanisms and function prediction of long noncoding RNAThe Scientific World Journal, 2012PMCID: PMC3540756
- Identification of novel candiyear maternal serum protein markers for Down syndrome by integrated proteomic and bioinformatic analysisPrenatal diagnosis, 2012PMID: 22430729
- A network-based gene-weighting approach for pathway analysisCell research, 2012PMID: 21894192
2011
- Comparative genomics study of polyhydroxyalkanoates (PHA) and ectoine relevant genes from Halomonas sp. TD01 revealed extensive horizontal gene transfer events and co-evolutionary relationshipsMicrobial cell factories, 2011PMID: 22040376
2010
- Lung adenocarcinoma from East Asian never-smokers is a disease largely defined by targetable oncogenic mutant kinasesJournal of clinical oncology : official journal of the American Society of Clinical Oncology, 2010PMID: 20855837
- FuncBase: a resource for quantitative gene function annotationBioinformatics, 2010PMCID: 2894510
2008
- Combining guilt-by-association and guilt-by-profiling to predict Saccharomyces cerevisiae gene functionGenome biology, 2008PMCID: 2447541
- An en masse phenotype and function prediction system for Mus musculusGenome biology, 2008PMCID: 2447542
- Isoform discovery by targeted cloning, ’deep-well’ pooling and parallel sequencingNature methods, 2008PMCID: 2743938
2006
- High precision multi-genome scale reannotation of enzyme function by EFICAzBMC genomics, 2006PMCID: 1764738
2004
- EFICAz: a comprehensive approach for accurate genome-scale enzyme function inferenceNucleic acids research, 2004PMCID: 535665
2003
- How well is enzyme function conserved as a function of pairwise sequence identity?Journal of molecular biology, 2003PMID: 14568541) (Faculty of 1000 Biology
2000
- Production of polyesters consisting of medium chain length 3-hydroxyalkanoic acids by Pseudomonas mendocina 0806 from various carbon sourcesAntonie van Leeuwenhoek, 2000PMID: 10696875
1999
- A rapid method for detecting bacterial polyhydroxyalkanoates in intact cells by Fourier transform infrared spectroscopyAppl Microbiol Biotechnol, 1999DOI:
1998
- Production of novel polyhydroxyalkanoates by Pseudomonas stutzeri 1317 from glucose and soybean oilFEMS Microbiol Lett, 1998DOI: